You are here
Science Resources: DNA Technologies
DNA Sequencing and What It Can Reveal About DNA Variation
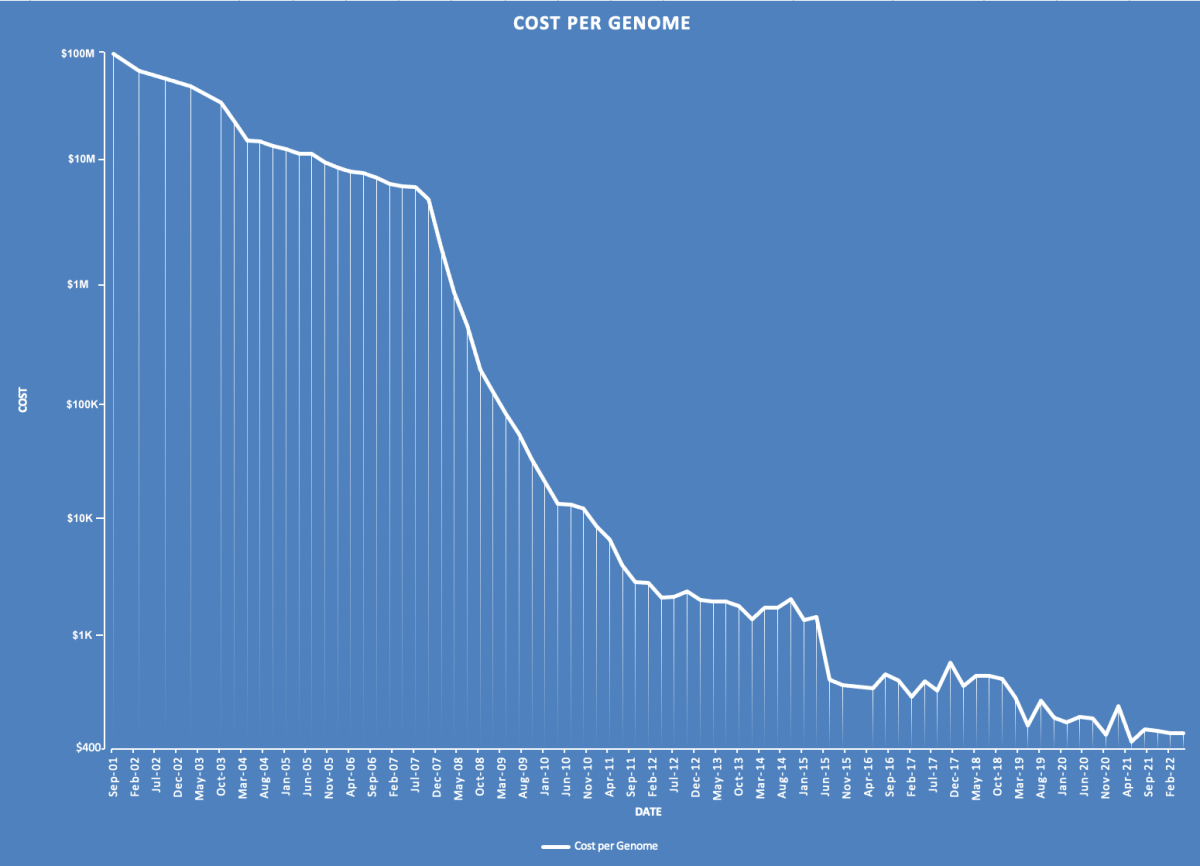
DNA sequencing is the process of reading every single base in a string of DNA. This can be performed for an individual’s entire genome or parts of their genome. The process of sequencing a whole genome has become dramatically cheaper and faster. As of 2023, commercial laboratories can sequence an individual’s entire genome as fast as a few hours and for just over $500, down from $100 million in the early 2000s (Fig. 5).[1]
Figure 5. The cost of sequencing a human genome has diminished rapidly in the past twenty years. The cost of sequencing a complete human genome has now reached approximately $500 and can be completed in less than a day.
Whole-Genome Sequencing
Whole-genome sequencing (WGS) reveals information about protein-coding, regulatory, and noncoding regions of the genome (Fig. 6). Sequencing the full human genome produces an immense amount of information. If you printed out a fully sequenced genome, the document would be over 250,000 pages long. The sheer volume of data generated by WGS and the fact that we still do not understand the functional importance of every single base in the genome or the impact of changes to any of those bases have limited the application of WGS for humans.
For the most part, full human genome sequencing is primarily used for research purposes and is not widely adopted for clinical or commercial uses. But as our ability to interpret the genome improves, there is a risk that whole genome sequences collected from individuals today could be exposed and subject to discrimination. Although there is some legislation on genetic nondiscrimination (the Genetic Information Nondiscrimination Act of 2008, or GINA)[2], how courts handle an exposure of such sensitive data will be a critical question going forward.
Figure 6. Sequencing can be applied at varying levels of the genome, either the whole genome, the exome, or only targeted regions. Determining the scope of a sequencing project is a balance between time, money, and data. Whole-genome sequencing can provide an abundance of data, but very little of it is yet interpretable. On the other hand, while targeted sequencing is fast and can provide actionable information, it won’t detect important variation outside the targeted regions.
Whole-Exome Sequencing
To avoid the high costs and immense data of whole-genome sequencing but still detect informative genetic variants, scientists commonly use an alternative method called whole exome sequencing (WES). This method sequences just the exome—the 2% of the genome that codes for proteins (Fig. 6). WES is typically much faster and more cost effective than whole-genome sequencing (~$200-$300 vs. $500). Because the exome is smaller, has a clear functional impact, and has been more extensively studied, it is easier to predict the effects of DNA variants in the exome.
For these reasons, WES is now an increasingly common tool in medical genetics, especially when attempting to diagnose a genetic disease caused by a mutation that is rare in the general population. The greater cost-effectiveness of WES has also made it more common among direct-to-consumer (DTC) personal genomic companies. DTC services may provide exome sequencing to customers along with interpretation of their genetic data and insights about ancestry, health, and personal attributes.
The courts are likely to see an increasing number of cases involving whole-exome sequencing as used in medical genetic testing. This also means they will face questions about the accuracy of detecting genetic variants and the interpretation of how these variants impact an individual’s health (see Algorithms Based on Genetic Data section).
Targeted or Gene Panel Sequencing
While the exome offers a narrower target than the whole genome, it may still be too broad and will also exclude information from noncoding regions of the genome. Targeted sequencing relies on predetermining specific genes or DNA regions to sequence, often as part of a gene panel (Fig. 6).
Targeted sequencing is useful when a single gene or set of DNA regions can be highly informative. For example, in cancer diagnosis and treatment, gene panels can be used to distinguish subtypes of a specific cancer (e.g., HER2+ vs. HER2- breast cancers).
Targeted sequencing can also be used as a surveillance tool. As shown in United States v. Casey, sequencing certain genes—like MT-CYB in the mitochondrial genome—can provide accurate species-level identification of a tissue sample that is otherwise unidentifiable, such as a filet of fish or canned crabmeat.[3]
When courts review cases involving targeted sequencing, along with questions of accuracy of variant detection and interpretation, they are also likely to consider if the specific genes sequenced are appropriate for the stated goal. For example, do they aid in accurate, species-level identification, or do they offer relevant and reliable information about a specified disease?
[1] Kris A. Wetterstrand, DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP), Nat’l Hum. Genome Resch. Inst., https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data (last updated May 16, 2023).
[2] Genetic Information Nondiscrimination Act of 2008, Pub. L. No. 110-233, 122 Stat. 881.
[3] United States v. Casey, No. 4:18cr4 (E.D. Va. filed Jan. 12, 2018).