You are here
Science Resources: DNA Technologies
DNA Genotyping: How It Differs from Sequencing and Relevant Methods
Unlike sequencing, where every nucleotide base of a DNA segment is read, genotyping determines the state of only a targeted set of genetic sites within the genome. Significant forethought is therefore required to determine which sites to genotype. Overall, less genetic data can be captured through genotyping than with sequencing, although the information that is captured can be incredibly informative about identity, health, ancestry, and behavior. Genotyping technologies are used broadly, including in forensic DNA identification, medicine, research, and for commercial purposes.
The benefit of genotyping technologies is that they are considerably faster and cheaper than sequencing. However, as sequencing costs decrease, genotyping arrays are slowly becoming obsolete, since the same information can be collected from short-read and long-read sequencing methods.
STR Genotyping
Most cases of forensic DNA identification involve DNA genotyping of short tandem repeats (STRs).[1] This method of forensic DNA identification is described in detail in the corresponding chapter of the Reference Manual on Scientific Evidence[2] and is briefly summarized here.
Short tandem repeats consist of a short motif of DNA, two to seven bases in length, that is repeated tens or hundreds of times. The exact number of repeats for a specific STR varies between individuals. There are numerous STRs within the human genome. Forensic identification databases have chosen a specific set of approximately 20 different STRs and use an individual’s combination of STR lengths as a highly specific “fingerprint” for identification. The individual’s identity is represented as a string of digits—like a serial number—representing the number of repeats at each of the genotyped STRs. Critically, the genotype at any of these STRs or the STRs in combination are not informative about the health, ancestry, or appearance of the individual.
SNP-Array Genotyping
Medical, research, and commercial genotyping often use a genotyping method that uses hundreds to millions of single nucleotide polymorphisms (SNPs)—sites in the genome where individuals differ by a single nucleotide. This technology is known as SNP-array or micro-array genotyping.[3]
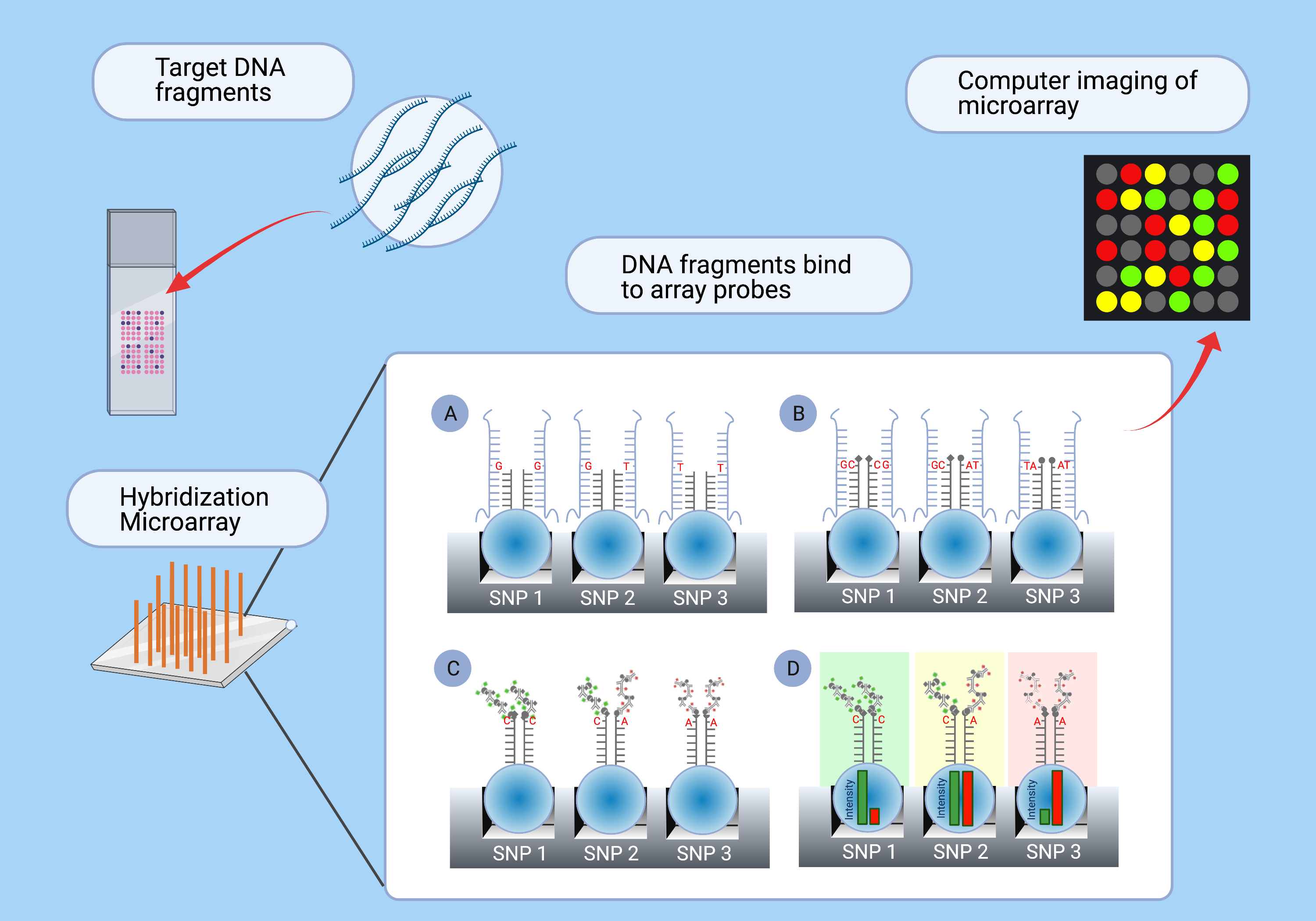
In SNP-array genotyping, molecular probes highly specific for certain sites in the genome are affixed to a silica chip. An individual’s DNA is made single-stranded, fragmented, and washed across the surface of the slide, binding to complementary probes (Fig. 11A). Fluorescently labeled nucleotides are added, which bind to the immobilized DNA strand at the site of the SNP (Fig. 11B–C), complementing the SNP nucleotide. The fluorescence is specific to the identity of the nucleotide base. A computer reads the fluorescence of each probe and determines the identity of the SNP (Fig. 11D).
Figure 11. Diagram of SNP-array genotyping. A microarray consists of DNA probes, targeting specific sites in the genome, affixed to a surface. The target DNA fragments are washed across the surface of the chip. A) The DNA fragments bind to the complementary probes. B) Fluorescently labeled nucleotides bind to the complementary SNPs in the DNA fragments. C) The DNA fragments are washed away, and the fluorescent labels are extended. D) The fluorescently labeled bases are imaged by a computer to determine the identity of the SNP.
In SNP genotyping, the molecular probes that bind the DNA are highly specific for certain sites in the genome. This means only a limited number of sites across the genome can be genotyped at a given time using a single array. There are an estimated 10 million commonly occurring SNPs across the human genome. A typical SNP-array chip may genotype between 500,000 and 2 million different SNPs. The SNPs to include on a given array are carefully considered before producing the array. SNP-arrays tend to be designed for specific purposes, like investigating SNPs associated with a particular disease or informing about genetic ancestry. The different purposes require choosing different SNPs to genotype.
The information obtained about an individual through SNP-array genotyping is substantially greater than what is gathered using forensic STR genotyping. The STRs chosen for forensic identification were picked explicitly to be uninformative about an individual’s health, ancestry, or behavior. SNP-array genotyping, however, can be highly informative about these features. Furthermore, it is possible to infer the identity of SNPs not genotyped on the array from those that were genotyped, through a process known as imputation. This means a remarkable amount of an individual’s genetic information can be determined using SNP-array genotyping.
Common Sources of Error in Genotyping
Similar sources of error are shared between sequencing and genotyping technologies.[4] For example, genotyping errors are often caused by the low quality and low quantity of DNA samples. DNA that is old, degraded, or exposed to chemical contaminants is likely to result in numerous errors during genotyping. Similarly, DNA samples that include contaminating DNA from other individuals or other organisms can produce inaccurate results. A small quantity of the DNA itself will also produce poor genotyping results.
In some cases, chemistry-based methods can be used to increase the amount of DNA in a sample before genotyping (PCR amplification). While generally an accurate process, these methods do have the potential to introduce artificial changes in the DNA sequence, producing errors during genotyping. There is also the opportunity for human error, such as sample mix-up, sample contamination, or mistakes during the genotyping process.
Common approaches to overcome genotyping errors involve adequate technician training, standardization of protocols, and regular internal validation. Additionally, regenotyping samples as technical replicates can catch potential errors and provide multiple reads of the same site in order to determine a consensus genotype. There is, however, a balance between cost and accuracy when deciding how many replicates are necessary to perform.
[1] John M. Butler, Short Tandem Repeat Typing Technologies Used in Human Identity Testing, 43 BioTechniques, Oct. 2007, at ii–v, https://doi.org/10.2144/000112582.
[2] David H. Kaye & George Sensabaugh, Reference Guide on DNA Identification Evidence, in Reference Manual on Scientific Evidence 129 (Nat’l Acad. of Sci. & Federal Judicial Center, 3d ed. 2011), available at https://www.fjc.gov/content/reference-manual-scientific-evidence-third-edition-1.
[3] Roger Bumgarner, Overview of DNA Microarrays: Types, Applications, and Their Future, 101 Current Protocols in Molecular Biology 22.1.1, 22.1.1–11 (Wiley 2013), available at https://doi.org/10.1002/0471142727.mb2201s101; David Gresham et al., Comparing Whole Genomes Using DNA Microarrays, 9 Nature Reviews Genetics 291 (2008), available at https://doi.org/10.1038/nrg2335.
[4] François Pompanon et al., Genotyping Errors: Causes, Consequences and Solutions, 6 Nature Reviews Genetics 847 (2005), available at https://doi.org/10.1038/nrg1707.