You are here
Science Resources: DNA Technologies
Determining Variant Pathogenicity and Enhanced Medical Testing
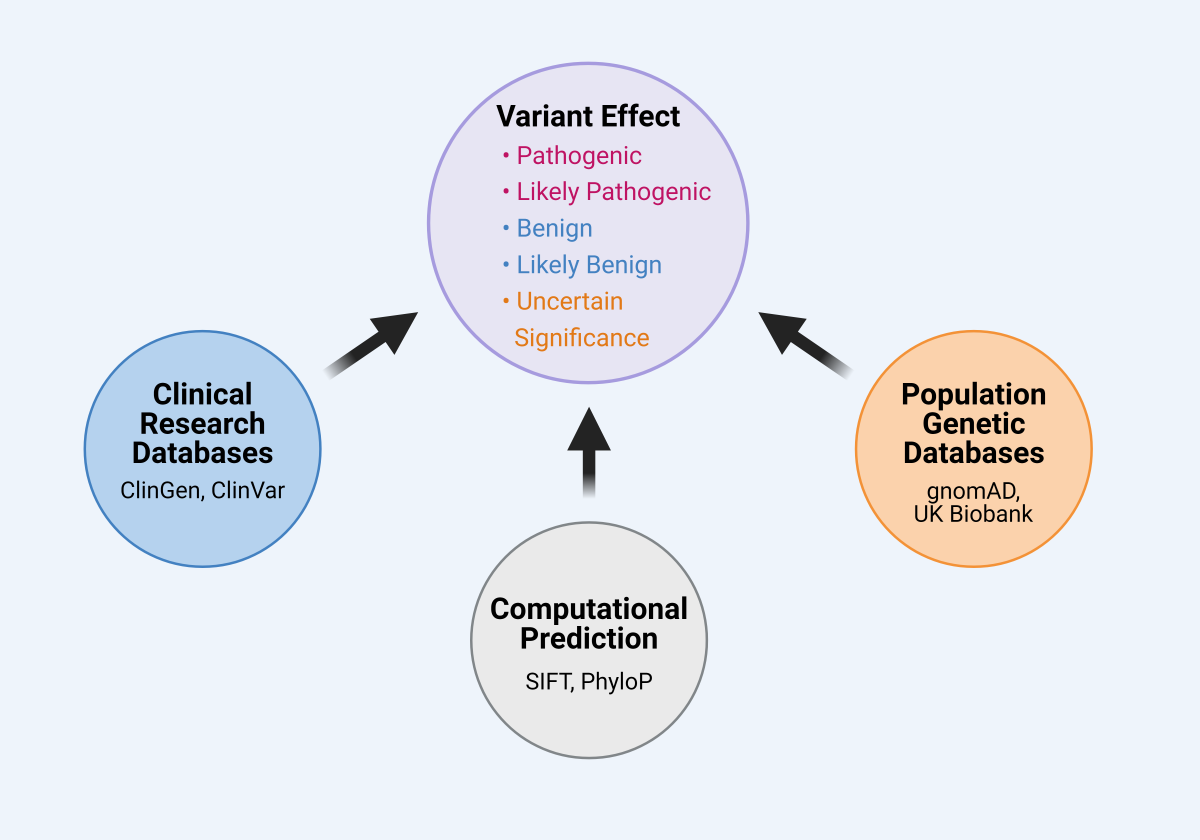
Classifying a genetic variant’s effect on human health relies on multiple sources of information (Fig. 18), and a variant’s classification may change over time as those sources are added to, reevaluated, or improved. Attributing effects to the millions of identified genetic variants is one of the critical hurdles in medical genetics and the burgeoning field of precision medicine. Often an individual’s genetic test reveals dozens of potential variants, for which only a subset have a documented effect. Variants detected through genetic testing are classified as the following:[1]
- benign or likely benign, indicating they are not known to cause disease
- pathogenic or likely pathogenic, indicating they directly contribute to an illness
- variant of uncertain/unknown significance (VUS)
Genetic testing laboratories classify a patient’s variants by reviewing the medical and genetics literature for reports linking a specific variant to a health outcome. This information is dispersed across scientific journals, clinicians’ reports, and individual researchers. Of course, studies and researchers may report contradictory findings.
Figure 18. Classifying a variant’s effect relies on multiples sources of information. A variant is classified as either pathogenic, likely pathogenic, benign, likely benign, or of uncertain significance. Classification relies on information from clinical databases (e.g., ClinGen, ClinVar), population genetic databases (e.g., gnomAD, UK Biobank), and computational approaches that predict the variant’s effect based on protein structure or evolutionary history (e.g., SIFT, PhyloP).
Databases and Tools for Genetic-Variant Classification
To facilitate the review of the research on genetic variants and their health effects, research and genetic testing communities have created centralized resources to collect and curate relevant data. Two such resources are ClinVar and the Clinical Genome Resource (ClinGen), both NIH-supported efforts that work in close partnership.[2]
ClinVar is a publicly searchable database that aggregates information about genetic variants and their relationship to human health by collecting submissions from genetic testing labs, researchers, and other databases.[3] ClinGen is an authoritative database that uses expert panels to curate submissions from clinicians, researchers, and patient groups and review published data for links between genetic variants and diseases.[4]
ClinVar may contain reports that conflict in their interpretation of a variant’s effects. However, ClinGen expert panels can review the data on these variants and submit standardized interpretations back to ClinVar as expert-reviewed records, resolving conflicting reports.
Genetic testing labs and clinicians often use ClinVar and ClinGen (and other databases) to consider which genetic variants to test for and which results to report to patients based on the evidence of a variant’s pathogenicity.
In addition to clinical studies, basic research has also been essential for classifying genetic variants. Large studies collecting genetic data from diverse populations have regularly contributed to variant reclassification.[5] For example, by studying variants that occur in the exomes of numerous diverse global populations, the Genome Aggregate Database (gnomAD) showed that many variants predicted to be pathogenic were found in exceptionally high numbers in certain populations, suggesting they are likely not pathogenic.[6]
Multiple computational tools are also available for predicting the pathogenicity of genetic variants,[7] combining information about how often a site in the genome differs across species and the likely effect a variant will have on a protein’s structure.[8]
Expert Guidelines for Classifying and Reporting Genetic Variants
There has also been a greater effort to normalize variant classification and reporting. Studies showed that testing labs often disagreed when classifying the same variant.[9] To improve agreement, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published guidelines for classifying variants.[10] These guidelines have been widely adopted and have enhanced agreement in classification among testing labs.[11]
Although the clinical implications of all laboratory results can change over time as knowledge advances, reexamining prior results is uncommon in medical care. Geneticists, however, are asking whether—and if so, when—prior results should be reclassified and returned to patients as new knowledge emerges. This is known as the “duty to recontact.”[12]
Possible triggers for reexamination include a request by the patient’s treating physician in light of new evidence or a laboratory’s decision to do so on its own. Reclassification can have important impact on patients’ health, such as altering treatment plans or easing emotional distress.
There is much debate about the extent to which reanalysis is ethically warranted or even required.[13] This issue is particularly challenging when laboratories initiate reclassification on their own, which may occur months or years after the initial genetic test. It may be difficult to return to patients results that permit effective care if patients have moved or changed healthcare providers.
The developing issues of genetic diagnoses and patient recontact also complicate cases of medical liability. Questions have arisen about the potential legal responsibility of laboratories and about statutes of limitations.[14] Since variant reinterpretation can occur repeatedly, it is unclear if the statute of limitation should apply to the date of the initial test, the reinterpretation, or the recontact. Determining the statute of limitations was an early point of contention in Williams v. Quest Diagnostics, Inc.[15] At issue was whether the period of time where Amy Williams could sue Athena began at the time of the initial diagnosis, or when she learned Athena had reclassified the variant.[16] Although so far, no courts have imposed liability on laboratories or physicians for failure to reexamine and return reclassified genomic results to patients,[17] healthcare providers’ potential liability in the future will depend on the extent that reclassification becomes routine.
[1] C. Sue Richards et al., ACMG Recommendations for Standards for Interpretation and Reporting of Sequence Variations: Revisions 2007, 10 Genetics in Med. 294 (2008), available at https://doi.org/10.1097/GIM.0b013e31816b5cae; Sue Richards et al., Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology, 17 Genetics in Med. 405 (2015) [hereinafter Richards et al., Joint Consensus], available at https://doi.org/10.1038/gim.2015.30.
[2] Melissa J. Landrum et al., ClinVar: Public Archive of Relationships Among Sequence Variation and Human Phenotype, 42 Nucleic Acids Resch. D980 (2014) [hereinafter Landrum et al., Public Archive], available at https://doi.org/10.1093/nar/gkt1113; Melissa J. Landrum et al., ClinVar: Improvements to Accessing Data, 48 Nucleic Acids Research D835 (2020), available at https://doi.org/10.1093/nar/gkz972; ClinVar and ClinGen Partnership, ClinGen, https://clinicalgenome.org/about/clingen-clinvar-collaboration/#:~:text=ClinGen (last visited Feb. 2, 2024); Heidi L. Rehm et al., ClinGen—The Clinical Genome Resource, 372 New Eng. J. Med. 2235 (2015), available at https://doi.org/10.1056/NEJMsr1406261.
[3] Landrum et al., Public Archive, supra note 2.
[4] Rhem et al., supra note 2.
[5] Rhem et al., supra note 2; Richards et al., Joint Consensus, supra note 1.
[6] Siwei Chen et al., A genomic mutational constraint map using variation in 76,156 human genomes,, 625 Nature 92 (2024), available at https://doi.org/10.1038/s41586-023-06045-0; Sanna Gudmundsson et al., Variant interpretation using population databases: Lessons from gnomAD, 43 Human Mutation 8 (2022), available at https://doi.org/10.1002/humu.24309.
[7] Katherine S. Pollard et al., Detection of Nonneutral Substitution Rates on Mammalian Phylogenies, 20 Genome Resch. 110 (2010), available at https://doi.org/10.1101/gr.097857.109.
[8] Pauline C. Ng & Steven Henikoff, SIFT: Predicting Amino Acid Changes That Affect Protein Function, 31 Nucleic Acids Resch. 3812 (2003), available at https://doi.org/10.1093/nar/gkg509; Robert Vaser et al., SIFT Missense Predictions for Genomes, 11 Nature Protocols 1 (2015), available at https://doi.org/10.1038/nprot.2015.123.
[9] Laura M. Amendola et al., Performance of ACMG-AMP Variant-Interpretation Guidelines Among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium, 98 Am. J. Hum. Genetics 1067 (2016), available at https://doi.org/10.1016/j.ajhg.2016.03.024.
[10] Richards et al., Joint Consensus, supra note 1.
[11] Annie Niehaus et al., A Survey Assessing Adoption of the ACMG-AMP Guidelines For Interpreting Sequence Variants and Identification of Areas for Continued Improvement, 21 Genetics in Med. 1699 (2019), available at https://doi.org/10.1038/s41436-018-0432-7.
[12] Karen L. David et al., Patient Re-Contact After Revision of Genomic Test Results: Points to Consider—A Statement of the American College of Medical Genetics and Genomics (ACMG), 21 Genetics in Med. 769 (2019), available at https://doi.org/10.1038/s41436-018-0391-z; Joshua L. Deignan et al., Points to Consider in the Reevaluation and Reanalysis of Genomic Test Results: A Statement of the American College of Medical Genetics and Genomics (ACMG), 21 Genetics in Medicine 1267 (2019), available at https://doi.org/10.1038/s41436-019-0478-1.
[13] Paul S. Appelbaum et al., Is There a Duty to Reinterpret Genetic Data? The Ethical Dimensions, 22 Genetics in Med. 633 (2020), available at https://doi.org/10.1038/s41436-019-0679-7; Deignan et al., supra note 12.
[14] Gary Marchant et al., Unjust Timing Limitations in Genetic Malpractice, 83 Albany L. Rev. 61 (2020), available at https://www.albanylawreview.org/article/70126; Alexandra L. Foulkes et al., Can Clinical Genetics Laboratories Be Sued for Medical Malpractice? 29 Annals Health L. 53 (2020), available at https://lawecommons.luc.edu/annals/vol29/iss1/5/.
[15] Williams v. Quest Diagnostics, Inc., 353 F. Supp. 3d 432 (D.S.C. 2018).
[16] See Turna Ray, Wrongful Death Suit Awaits Input from South Carolina Supreme Court, GenomeWeb (Apr. 4, 2017), https://www.genomeweb.com/molecular-diagnostics/wrongful-death-suit-awaits-input-south-carolina-supreme-court#.YNPqrpNKg8M; South Carolina Supreme Court Decision Deals Blow to Plaintiff in Quest Wrongful Death Suit, GenomeWeb (June 28, 2018), https://www.360dx.com/clinical-lab-management/south-carolina-supreme-court-decision-deals-blow-plaintiff-quest-wrongful#.YNPr4pNKg8N.
[17] See Ellen Wright Clayton et al., Does the Law Require Reinterpretation and Return of Revised Genomic Results?, 23 Genetics in Med. 833 (2021), available at https://doi.org/10.1038/s41436-020-01065-x.