You are here
Science Resources: DNA Technologies
Genomic Reference Databases Catalogue Human Genetic Variation
Fast and cheap genome sequencing has facilitated the collection of large genetic research databases. These genetic databases may contain little additional nongenetic information about participants, or they may link genetic data with information about health, appearance, and behavior. Generally, these data have had direct identifiers removed and are provided based on researchers’ promise (e.g., via a data-use agreement) that they will not attempt to reidentify participants. In general, these databases carry a low risk to participants for reidentification, exposure, and discrimination. But reviewing the details of these databases is useful for later discussions about genetic interpretation tools and algorithms, which rely heavily on these databases.
Population Reference Databases Support the Study of Genetic Variation and Common Traits
Catalogs of global human genetic variation have been critical to studies of human history and common genetic diseases (e.g., diabetes, cancer, stroke, depression). Some examples include the 1000 Genomes Project (1kGP), based on whole-genome sequence data;[1] the Simons Genome Diversity Project (SGDP), also based on whole-genome sequence data,[2] and the Genome Aggregation Database (gnomAD), based on whole-genome and whole-exome sequence data.[3]
These databases contain information about globally diverse populations, broadly capturing the genetic variation present in African, European, East Asian, South Asian, and Indigenous American populations (Fig. 12). Other studies aim to map the genetics of geographic and ethnic subgroups, for example, the African Genome Variation Project, which collected genotype and whole-genome sequence data from individuals across sub-Saharan Africa.[4]
Figure 12. Schematic of global collection for genetic reference databases. Population reference databases collect the genetic information of individuals from globally diverse populations. While populations share a great amount of genetic variation, some variants are shared by only a few populations, or are specific to a population. These patterns of shared variation are shaped over the past several millennia by histories of migration, population mixture, and genetic adaptations to local environments.
Aside from genetic data and limited information about individuals’ geographic origin, sex, and ethnicity, these reference databases contain no additional personal identifiable information about individuals or additional information about their health, appearance, or behavior. The limited amount of identifiable information lessens concerns about participant reidentification.
Participants who have donated genetic data to these databases have given consent for their information to be used by multiple future studies. Participants are not asked to consent to non-research use, such as access for marketing or law enforcement purposes. The scope of future studies is typically constrained—albeit to a broad range of topics (e.g., health, evolution, population history) and a broad range of researchers.
Access to these data is often freely available, and the data can be downloaded directly from the database webpage by anyone. The only barrier to accessing these data is often whether they have yet been published in a scientific journal.
Genome-Wide Association Studies Link Genetics to Complex Traits
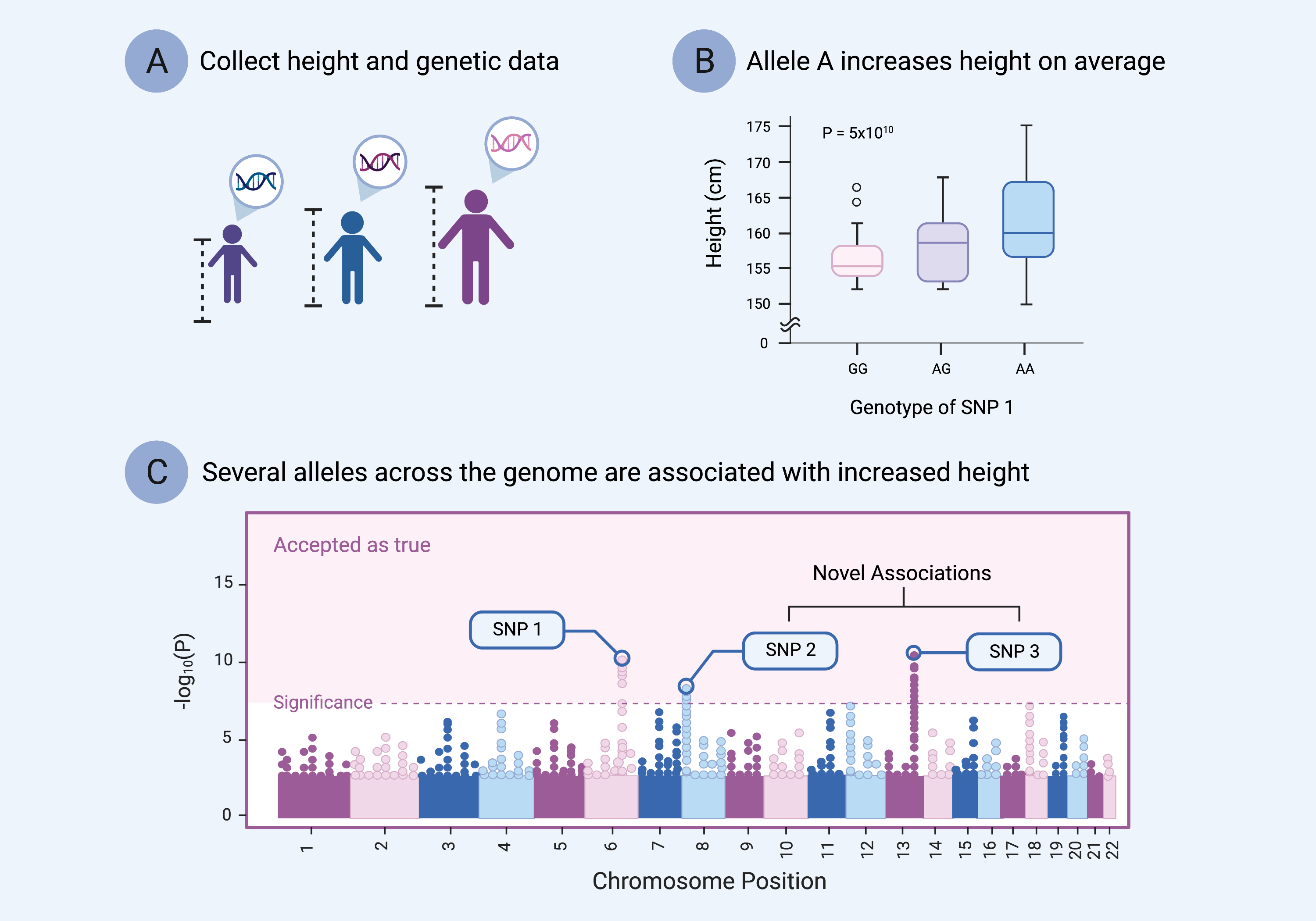
The best way to understand the genetic basis of a trait is to compare genetic data from a group of people with that trait to genetic data from a group of people without that trait. This approach has led to the common genetic study design known as a genome-wide association study (GWAS) (pronounced “gee-was”) (Fig. 13).[5]
GWAS are the cornerstone approach for studying common and complex genetic traits—where many genetic variants each contribute a small effect to the overall trait—such as height, heart disease, or smoking behavior.[6] The basic GWAS design follows three steps:
- Recruit individuals with and without a particular trait for discrete traits (e.g., schizophrenia), or that fall along a spectrum of a continuous trait (e.g., height) (Fig. 13A).
- Collect detailed genetic information from those individuals (usually genome-wide SNP data) along with detailed information about the trait or traits of interest (Fig. 13A).
- Perform statistical analyses to determine if a specific genetic variant (fig. 13B) or set of variants (fig. 13C) are more often associated with the trait of interest than not.[7]
Figure 13. Principles of a Genome-Wide Association Study. A) Trait and genetic data are collected for a large cohort of individuals. The genetic data are most often genome-wide SNP data. B) Statistical analyses are used to identify a variant or set of variants associated with differences in the trait of interest. C) Variants across the genome are tested for their association to the trait of interest, and a strict statistical threshold is used to prevent false discoveries. The strength of the variants’ associations with the trait are depicted as a Manhattan plot.
GWAS require the analysis of genetic data from thousands of individuals, so participants are often asked to give consent to allow GWAS datasets to be used across multiple studies by different researchers and to be combined in meta-analyses, although studies are usually constrained to health and disease research. The transfer of data across research groups and the link between genetic and trait data increases the risk to participants of reidentification even from data for which direct identifiers have been removed.
Because of this, institutional review boards (IRBs), national health services, and the researchers’ discretion act as the gatekeepers to determine who can access the data and for what purpose. Technical approaches that alter the DNA data in ways that frustrate reidentification but do not hamper scientific analyses have also been considered but are not widely used.[8]
About 80% of the genetic data collected through GWAS so far are from individuals with European ancestry.[9] This bias in genetic data limits the ability to make new discoveries and genetic tools, and to extend these discoveries and tools to non-European populations.[10] Efforts are underway to rectify the lack of diversity in genetic datasets, with studies like Trans-Omics for Precision Medicine (TOPMed) and Population Architecture using Genomics and Epidemiology (PAGE) collecting genetic and clinical data from ethnically diverse populations.[11]
[1] The 1000 Genomes Project Consortium, A Global Reference for Human Genetic Variation, 526 Nature 68 (2015), available at https://doi.org/10.1038/nature15393.
[2] Swapan Mallick et al., The Simons Genome Diversity Project: 300 Genomes from 142 Diverse Populations, 538 Nature 201 (2016), available at https://doi.org/10.1038/nature18964.
[3] Konrad J. Karczewski et al., The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans, 581 Nature 434 (2020), available at https://doi.org/10.1038/s41586-020-2308-7.
[4] Deepti Gurdasani et al., The African Genome Variation Project Shapes Medical Genetics in Africa, 517 Nature 327 (2014), available at https://doi.org/10.1038/nature13997.
[5] Nat’l Hum. Genome Resch. Inst., Genome-Wide Association Studies Fact Sheet [hereinafter NHGRI Fact Sheet], https://www.genome.gov/about-genomics/fact-sheets/Genome-Wide-Association-Studies-Fact-Sheet (last updated Aug. 17, 2020); Jacqueline MacArthur et al., The New NHGRI-EBI Catalog of Published Genome-Wide Association Studies (GWAS Catalog), 45 Nucleic Acids Resch. D896 (2017), available at https://doi.org/10.1093/nar/gkw1133; Peter M. Visscher et al., Five Years of GWAS Discovery, 90 Am. J. of Hum. Genetics, Jan. 13, 2012, at 7–24, available at https://doi.org/10.1016/j.ajhg.2011.11.029.
[6] MacArthur et al., supra note 5.
[7] NHGRI Fact Sheet, supra note 5.
[8] Yaniv Erlich, Y., & Arvind Narayanan, Routes for Breaching and Protecting Genetic Privacy, 15 Nature Reviews Genetics 409 (2014), available at https://doi.org/10.1038/nrg3723.
[9] Slavé Petrovski & David B. Goldstein, Unequal Representation of Genetic Variation Across Ancestry Groups Creates Healthcare Inequality in the Application of Precision Medicine, 17 Genome Biology, no. 157, at 1–3 (2016), available at https://doi.org/10.1186/s13059-016-1016-y; Alice B. Popejoy & Stephanie M. Fullerton, Genomics Is Failing on Diversity, 538 Nature 161 (2016), available at https://doi.org/10.1038/538161a.
[10] L. Duncan et al., Analysis of Polygenic Risk Score Usage and Performance in Diverse Human Populations, 10 Nature Commc’ns, July 25, 2019, at 1–9, available at https://doi.org/10.1038/s41467-019-11112-0; Genevieve L. Wojcik et al., Genetic Analyses of Diverse Populations Improves Discovery for Complex Traits, 570 Nature 514 (2019), available at https://doi.org/10.1038/s41586-019-1310-4.
[11] About TOPMed, NHLBI Trans-Omics for Precision Med. WGS, https://www.nhlbiwgs.org/ (last updated Nov. 30, 2020); Home, The PAGE Study: Population Architecture Using Genomics & Epidemiology, https://www.pagestudy.org/ (last visited June 21, 2021).